Google AI, in collaboration with the UC Santa Cruz Genomics Institute, has launched DeepPolisher, a cutting-edge deep studying device designed to considerably enhance the accuracy of genome assemblies by correcting base-level errors. Its notable efficacy was lately demonstrated in advancing the Human Pangenome Reference, a serious milestone in genomics analysis.
The Problem of Correct Genome Meeting
A reference genome is a vital basis for understanding genetic variety, heredity, illness mechanisms, and evolutionary biology. Fashionable sequencing applied sciences, together with these developed by Illumina and Pacific Biosciences, have dramatically improved sequencing accuracy and throughput—however even with technological breakthroughs, assembling an error-free human genome (comprising over 3 billion nucleotides) stays immensely difficult. Even a minuscule per-base error fee may end up in hundreds of errors which might obscure key genetic variations or mislead downstream analyses.
What Is DeepPolisher?
DeepPolisher is an open-source, transformer-based sequencing correction device. Constructing on advances from DeepConsensus, it takes benefit of transformer deep studying architectures to additional cut back errors in genome meeting, notably insertion and deletion (indel) errors, which have a profound affect by shifting studying frames and may trigger necessary genes or regulatory parts to be missed throughout annotation.
- Expertise: Encoder-only transformer, adapting confirmed strategies in pure language processing for genomics.
- Coaching information: Leveraged a human cell line extensively characterised by NIST and NHGRI, sequenced with numerous platforms to make sure near-complete accuracy (~99.99999% correctness, between 300–1,000 errors in 6 billion bases).
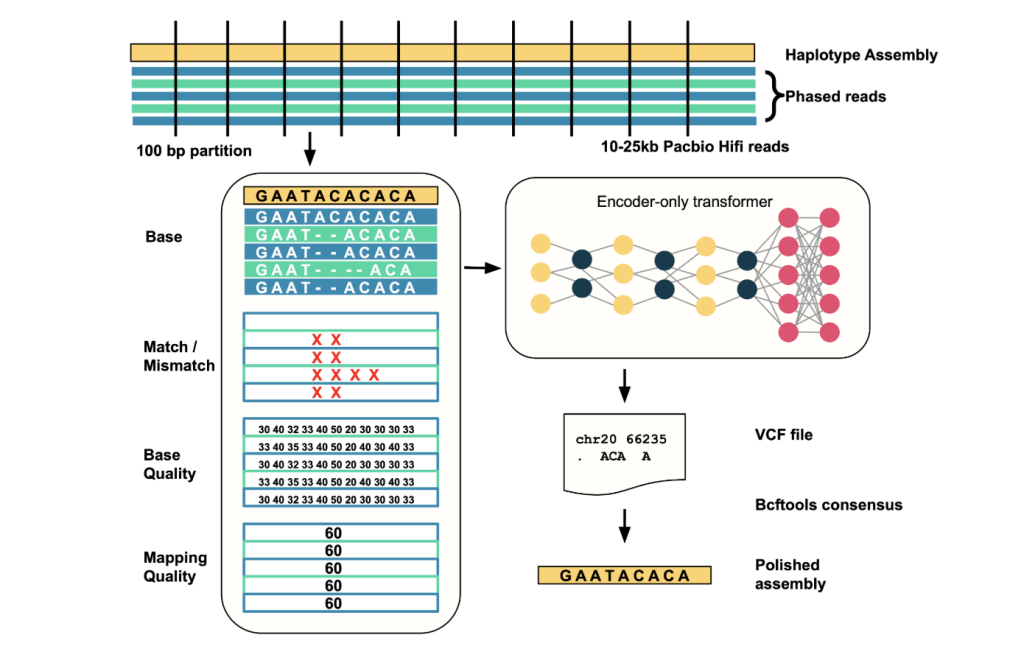
How Does It Work? (Technical Overview)
- Enter Alignment: Takes aligned PacBio HiFi reads towards a haplotype-resolved genome meeting as enter.
- Error Website Detection: Scans the meeting in 25kb home windows; identifies candidate error websites the place learn proof deviates from the meeting.
- Knowledge Encoding: For every window containing putative errors (<100bp), it creates a multi-channel tensor illustration of learn alignment options similar to base, base high quality, mapping high quality, and match/mismatch standing.
- Mannequin Inference: Feeds these tensors into the transformer, which predicts corrected sequences for these areas.
- Output Correction: Outputs variations in VCF format, that are then utilized to the meeting to provide a cultured, extremely correct sequence utilizing instruments like bcftools.
Efficiency and Impression
DeepPolisher delivers substantial enhancements:
- Whole error discount: ~50%
- Indel error discount: >70%
- Error charges: Achieves an error fee as little as one base error per 500,000 assembled bases in real-world deployment with the Human Pangenome Reference Consortium (HPRC).
- Genomic Q-score enchancment: Raises meeting high quality from Q66.7 to Q70.1 on common (Q-score is a logarithmic measure of per-base error fee; increased is best. Q70.1 implies <1 error per 12 million nucleotides)
- Each pattern examined by HPRC confirmed enchancment.
These advances immediately affect the reliability and accuracy of derived references, similar to within the Human Pangenome Reference, which noticed a fivefold information enlargement and substantial error discount on account of DeepPolisher.


Deployment and Functions
- Built-in in main tasks: Utilized in HPRC’s second information launch, offering high-accuracy reference assemblies for 232 people, making certain broad ancestral variety in genomic references.
- Open-source entry: Obtainable by way of GitHub, with case research and Dockerized workflows to be used on assemblies produced by instruments like HiFiasm and sequenced with PacBio HiFi reads.
- Generalizability: Whereas initially centered on human genomes, the construction and method are adaptable to different organisms and sequencing platforms, fostering accuracy throughout the genomics group.
Sensible Workflow Instance
A typical workflow utilizing DeepPolisher would possibly contain:
- Enter: HiFiasm diploid meeting and PacBio HiFi reads, phase-aligned utilizing the PHARAOH pipeline.
- Working: Dockerized instructions for picture creation, inference, and correction software.
- Output: Separate VCF recordsdata for maternal and paternal assemblies, polished FASTAs after bcftools consensus step.
- Evaluation: Use of benchmarking instruments (e.g., dipcall, Hap.py) to quantify enhancements in error charges and variant accuracy.
Conclusion and Future Instructions
DeepPolisher represents a leap ahead in genome sprucing know-how—sharply decreasing error charges and unlocking increased decision for practical genomics, uncommon variant discovery, and scientific functions. By concentrating on the remaining barrier to good genome assemblages, it permits extra correct prognosis, population-level genetic research, and paves the way in which for next-generation reference tasks benefiting biomedical analysis and drugs.
Try the Technical particulars, GitHub Web page and Paper. Be at liberty to take a look at our GitHub Web page for Tutorials, Codes and Notebooks. Additionally, be at liberty to comply with us on Twitter and don’t neglect to affix our 100k+ ML SubReddit and Subscribe to our E-newsletter.
Asif Razzaq is the CEO of Marktechpost Media Inc.. As a visionary entrepreneur and engineer, Asif is dedicated to harnessing the potential of Synthetic Intelligence for social good. His most up-to-date endeavor is the launch of an Synthetic Intelligence Media Platform, Marktechpost, which stands out for its in-depth protection of machine studying and deep studying information that’s each technically sound and simply comprehensible by a large viewers. The platform boasts of over 2 million month-to-month views, illustrating its reputation amongst audiences.


